Build living tissue simulations with the Active-Finite-Voronoi model.
PyAFV is a simple Python toolkit for generating 2D finite-Voronoi tessellations, modeling cell mechanics, and exploring collective motility under active dynamics.
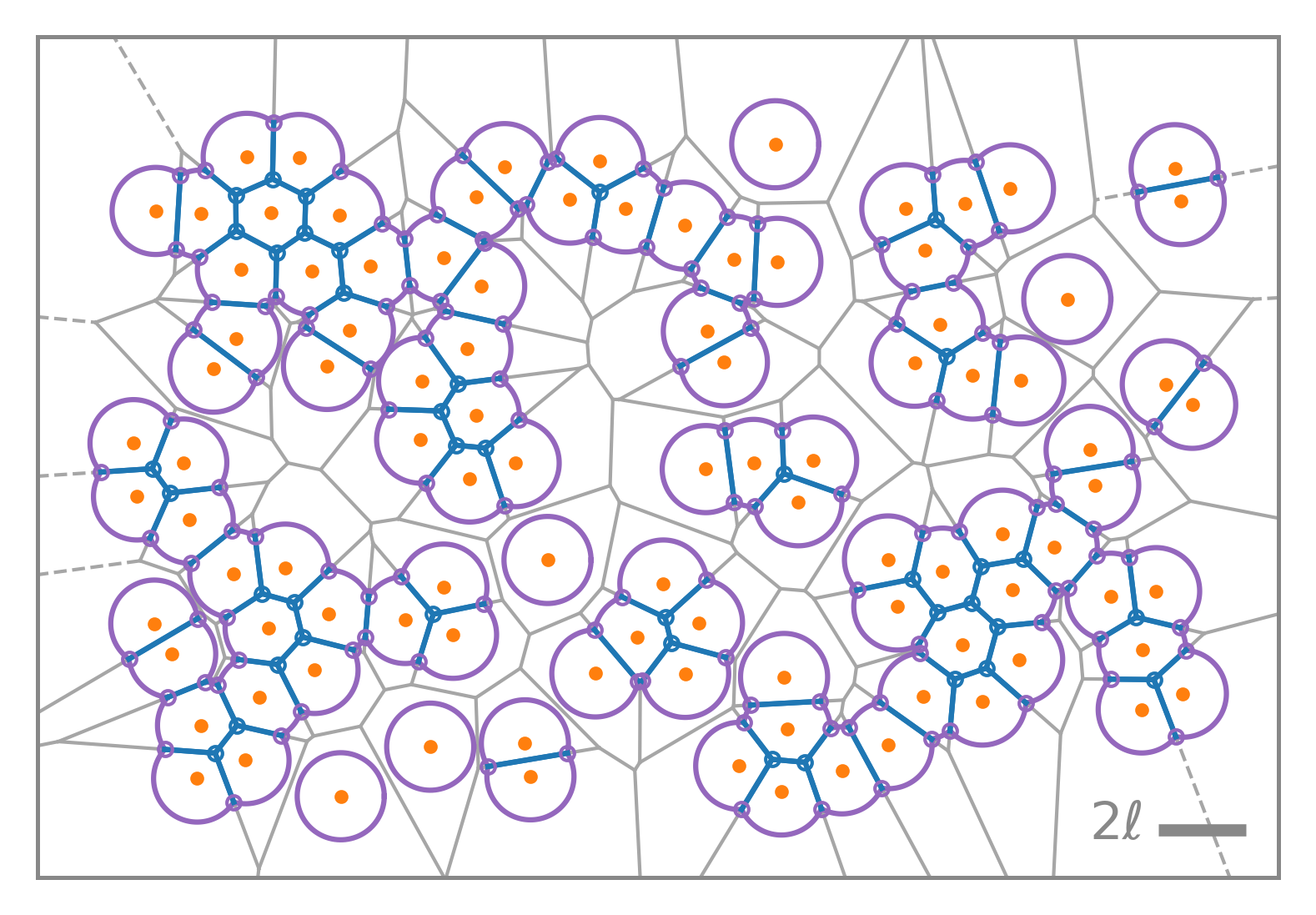
Geometric insight, physical fidelity
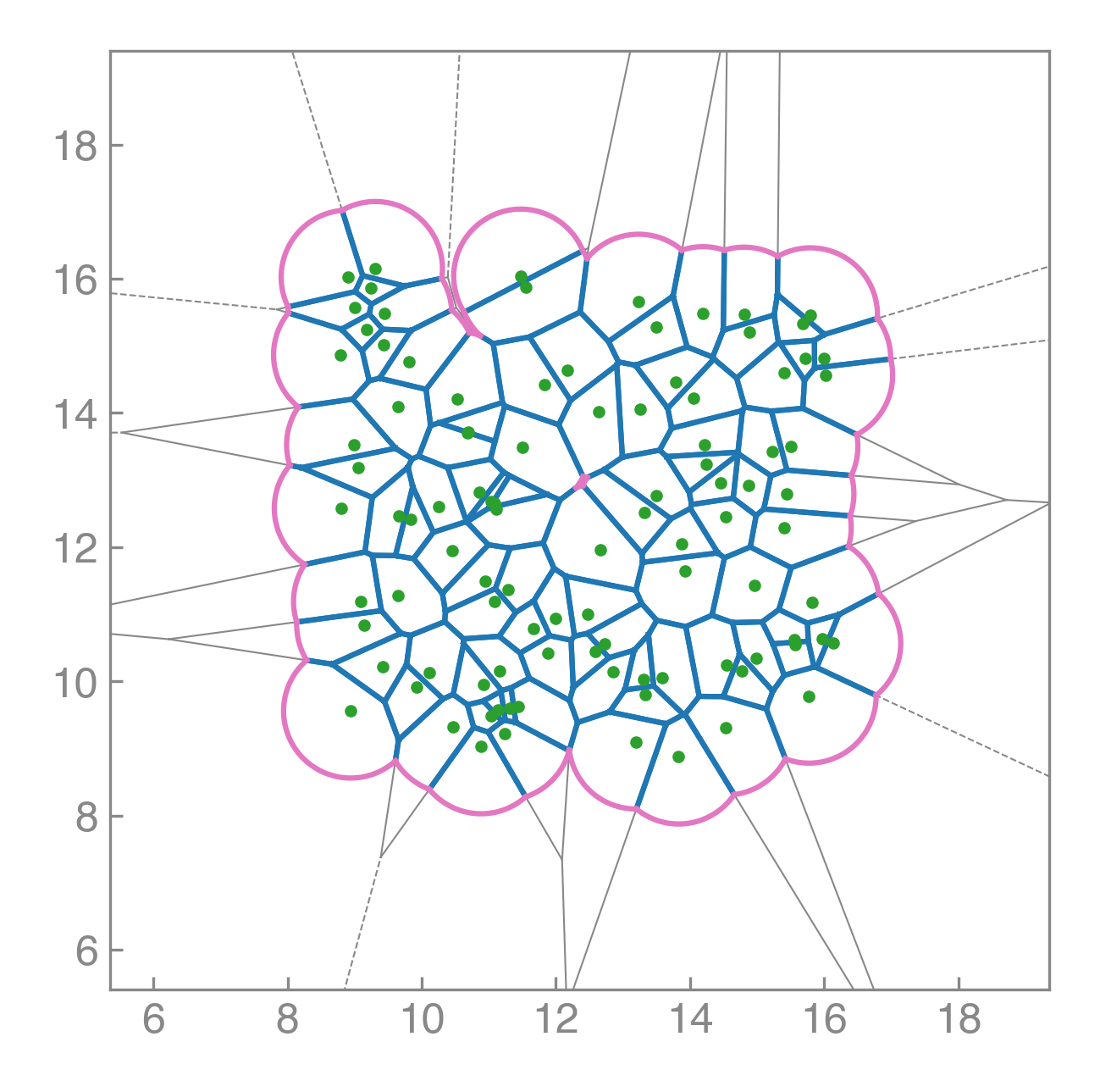
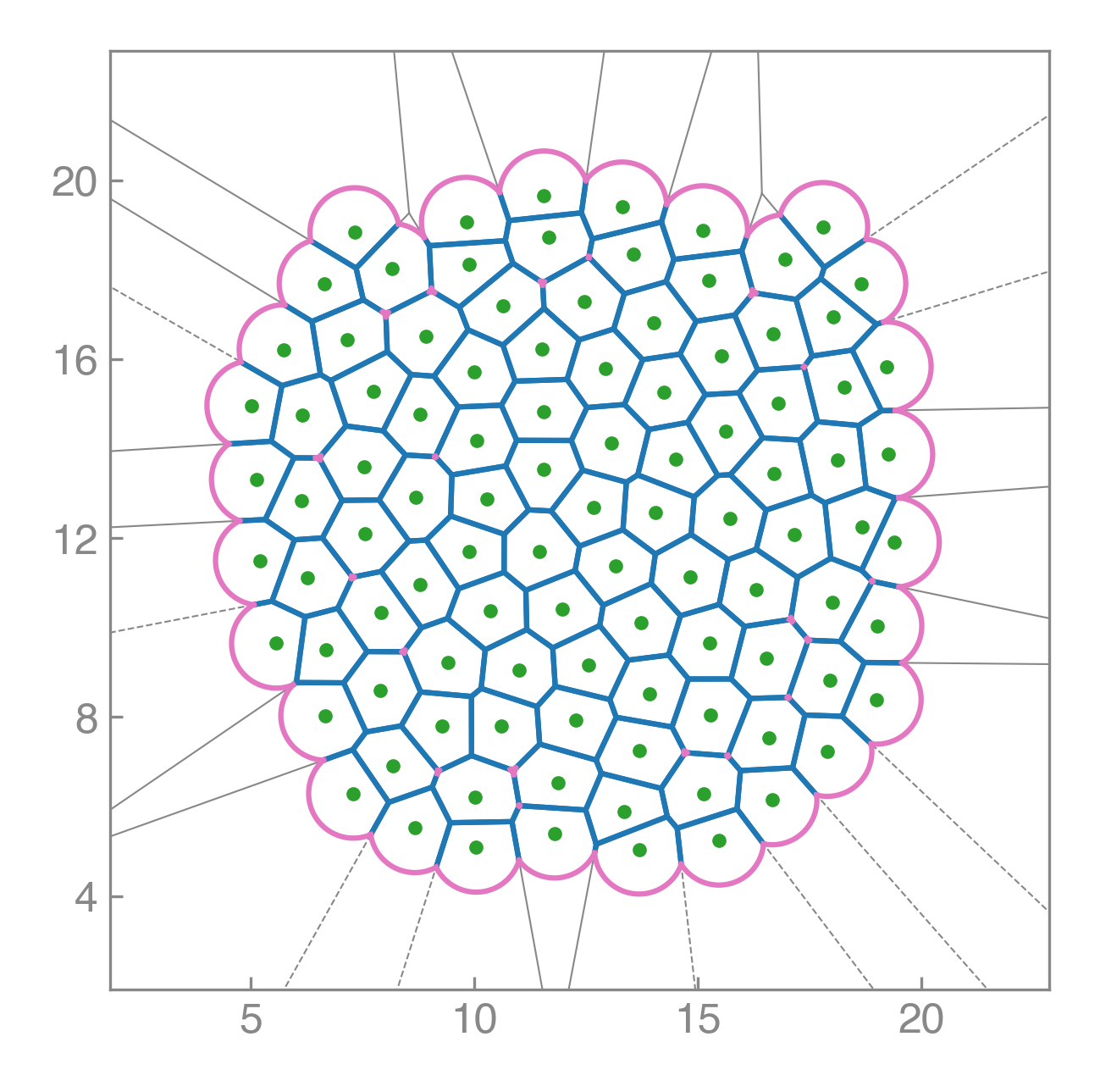
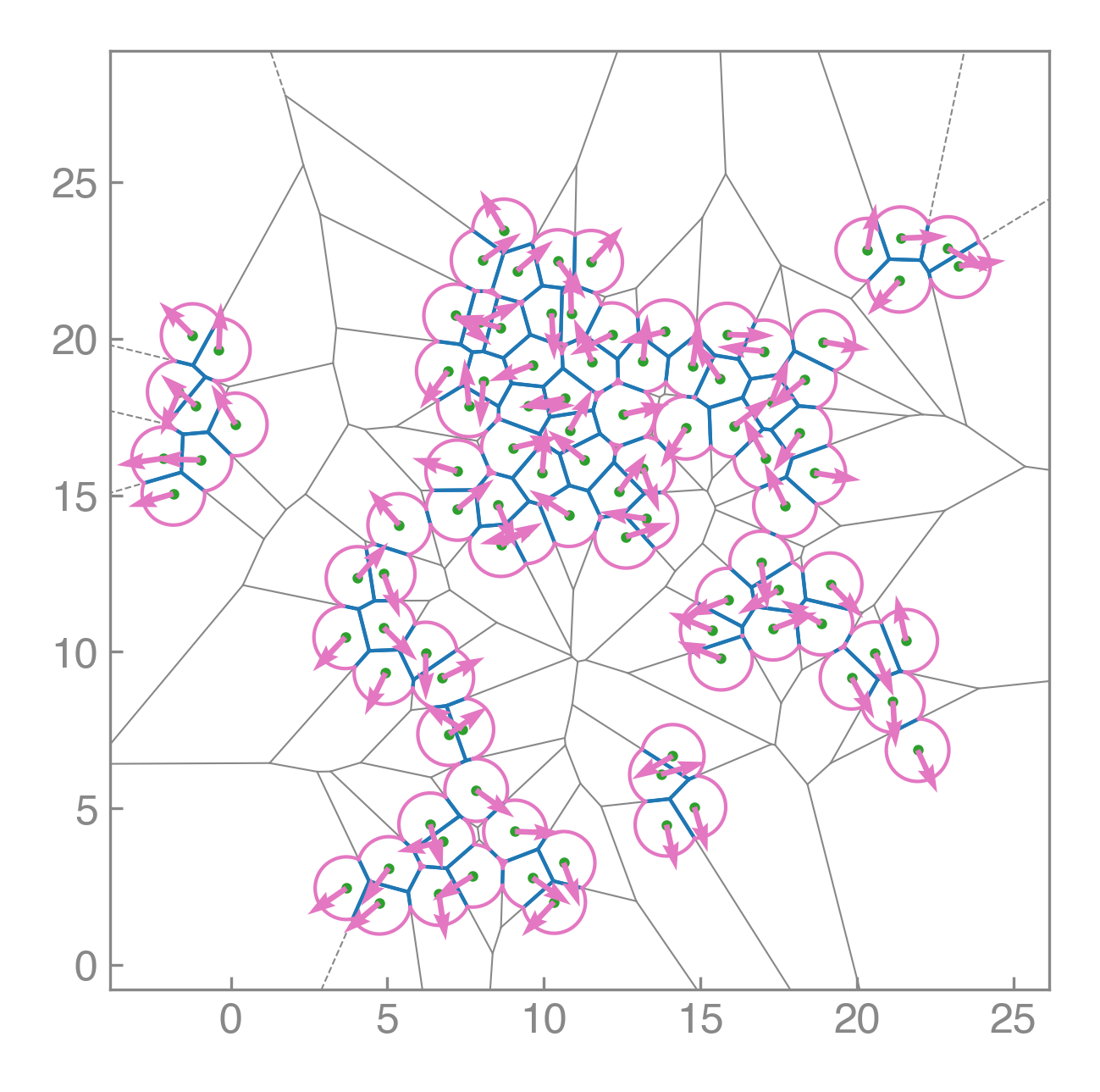
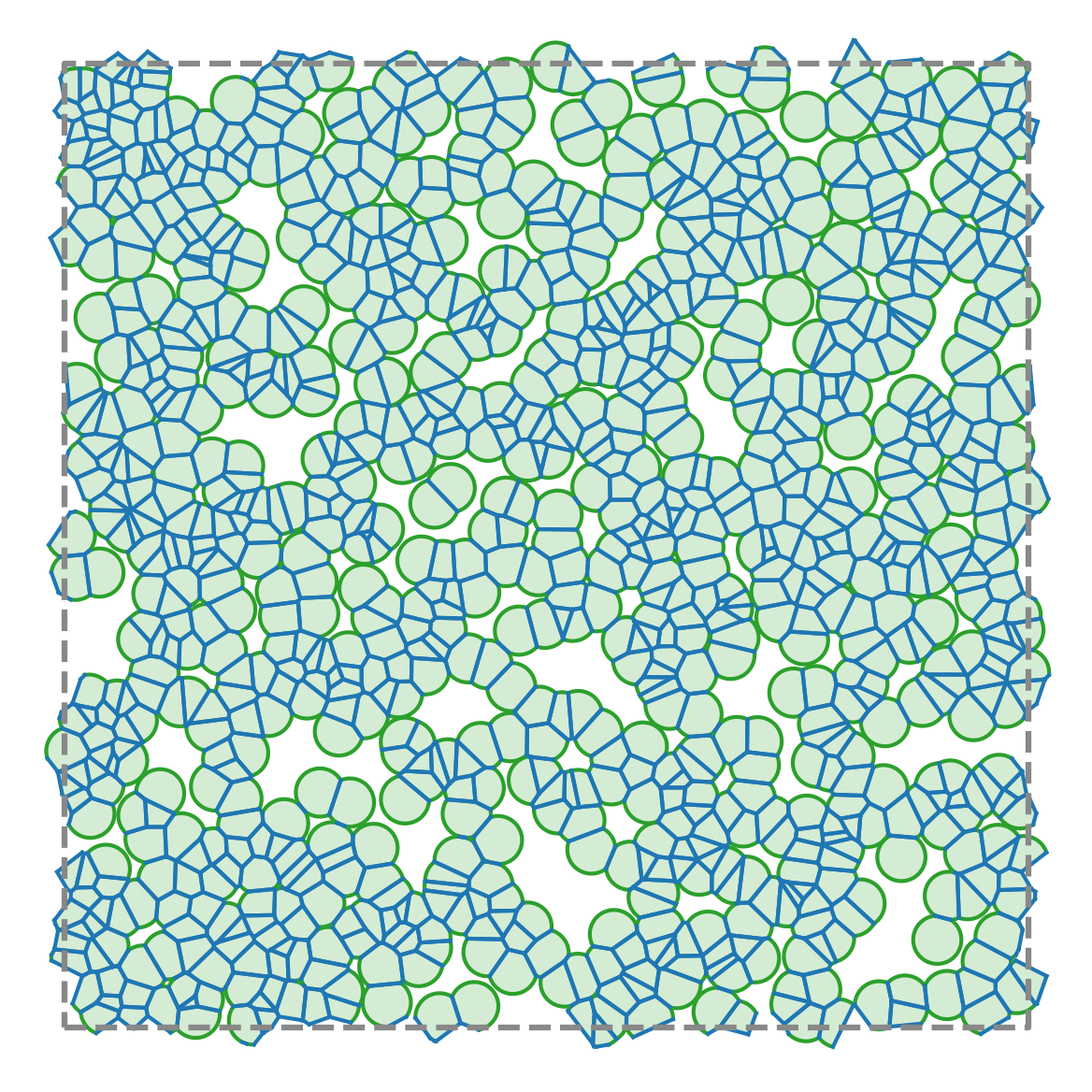
Compute areas, perimeters, vertices, edges, and conservative forces while visualizing emergent tissue structure in real time.